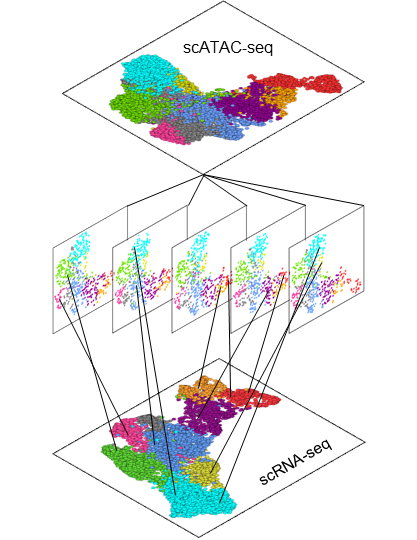
 Single Cell (sc) Analysis Unit is a new core facility initiative with the mission to provide analysis services and technical support to researchers wanting to conduct single cell Next Generation Sequencing (scNGS) studies. We offer our expertise and computational space to support our clients’ research by providing analysis services for a wide range of single cell assays including scRNA-seq (single cell profiling of the transcriptome), scATAC-seq (single cell profiling of the epigenome), and spatial transcriptomics (sequencing-based spatial profiling and imaging-based in situ spatial profiling). The Single Cell Analysis Unit collaborates closely with the Greek Research Infrastructure for Personalised Medicine – pMedGR for Single Cell analysis and NGS, ensuring integrated and high-quality workflows (https://www.precisionmedicine.gr/).
Single Cell (sc) Analysis Unit is a new core facility initiative with the mission to provide analysis services and technical support to researchers wanting to conduct single cell Next Generation Sequencing (scNGS) studies. We offer our expertise and computational space to support our clients’ research by providing analysis services for a wide range of single cell assays including scRNA-seq (single cell profiling of the transcriptome), scATAC-seq (single cell profiling of the epigenome), and spatial transcriptomics (sequencing-based spatial profiling and imaging-based in situ spatial profiling). The Single Cell Analysis Unit collaborates closely with the Greek Research Infrastructure for Personalised Medicine – pMedGR for Single Cell analysis and NGS, ensuring integrated and high-quality workflows (https://www.precisionmedicine.gr/).
Our team offers end-to-end single-cell analysis solutions, using state-of-the art bioinformatics methodologies and in-house analysis pipelines, in order to offer structured, high quality and reproducible analysis deliverables. We also want to guide our community to have easy access to primary analysis solutions, and we aim to develop novel algorithms that will interpret the high-dimensional single-cell information in order to answer complex biological questions.
The Unit has developed SCALA (Single Cell AnaLysis for All), an R/Shiny web application and stand-alone toolkit, that handles the analysis of scRNA-seq and scATAC-seq datasets. It offers various modes of analysis including quality control, normalization, dimensionality reduction, differential expression/accessibility analysis, cell clustering, functional enrichment analysis, trajectory inference, cell-to-cell communication analysis, gene regulatory network (GRN) reconstruction, and visualization. For more information and to try out the web version of the tool, please visit - http://scala.fleming.gr/app/scala , or visit - https://github.com/PavlopoulosLab/SCALA to install it locally.
Furthermore, robust workflows for the analysis, exploration, and interpretation of single-cell and spatially resolved transcriptomics data have been established by the unit, supporting a range of biological projects across multiple tissues and disease contexts, including rheumatoid arthritis (RA), multiple sclerosis (MS), inflammatory bowel disease (IBD) and COVID-19. Objectives that have been successfully completed include comparative analyses between healthy and diseased samples at various levels — such as differentially expressed genes (DEGs), enriched functional terms, gene regulatory networks (GRNs), and cell-to-cell communication patterns. Additionally, integration across modalities such as RNA and ATAC has been carried out, along with the deconvolution of spatial transcriptomics data using matched single-cell RNA-seq datasets as reference.
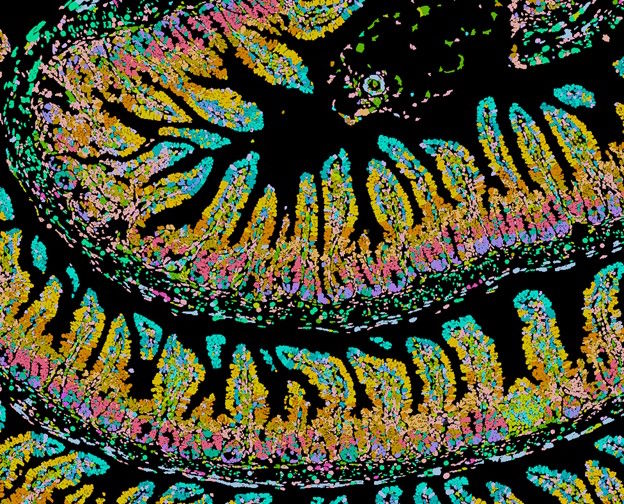
Recently, our unit has expanded its research into the emerging field of spatial transcriptomics, which enables the investigation of gene expression within the spatial context of tissues. Among the various technological platforms available, we have analyzed data from both the high-throughput sequencing-based Visium 10X and the image-based, single-cell resolution Xenium 10X systems. These tools allow us to characterize cellular neighborhoods, delineate broader anatomical regions, and explore cell–cell interactions in a spatially informed framework. When applied to case-control samples, our workflow can be extended to detect condition-specific spatial variations that may influence the onset and progression of the phenotype under study.
For more information or to request a service, please click here.
Unit development is supported by the Hellenic Foundation for Research & Innovation (HFRI) ELIDEK; 1st Call for H.F.R.I. Research Projects to Support Faculty Members & Researchers and Procure High-Value Research Equipment (Grant ID: 3780) to G. Kollias.
Publications
Armaka M, Konstantopoulos D, Tzaferis C, Lavigne MD, Sakkou M, Liakos A, Sfikakis PP, Dimopoulos MA, Fousteri M & Kollias G. 2022, "Single-cell multimodal analysis identifies common regulatory programs in synovial fibroblasts of rheumatoid arthritis patients and modeled TNF-driven arthritis", Genome Med. , 14(1) :78. [PubMed].
Tzaferis C, Karatzas E, Baltoumas FA, Pavlopoulos GA, Kollias G, Konstantopoulos D. 2023, SCALA: A complete solution for multimodal analysis of single-cell Next Generation Sequencing data. Comput Struct Biotechnol J. , 21:5382-5393. [PubMed].
Roumelioti F, Tzaferis C, Konstantopoulos D, Papadopoulou D, Prados A, Sakkou M, Liakos A, Chouvardas P, Meletakos T, Pandis Y, Karagianni N, Denis MC, Fousteri M, Armaka M, Kollias G. 2024, Mir221/222 drive synovial hyperplasia and arthritis by targeting cell cycle inhibitors and chromatin remodeling components. Elife. , 5;13:e84698. [PubMed].
Iliopoulou L, Tzaferis C, Prados A, Roumelioti F, Koliaraki V, Kollias G. 2025, Different fibroblast subtypes propel spatially defined ileal inflammation through TNFR1 signalling in murine ileitis. Nat Commun. , 16(1):3023. [PubMed]
Detsika MG, Sakkou M, Triantafyllidou V, Konstantopoulos D, Grigoriou E, Psarra K, Jahaj E, Dimopoulou I, Orfanos SE, Tsirogianni A, Kollias G, Kotanidou A. 2025, CD55 upregulation in T cells of COVID-19 patients suppresses type-I interferon responses. Commun Biol. , 2;8(1):690. [PubMed]
Unit Members
Christos Tzaferis / Postdoctoral Fellow
Konstantinos Apostolou-Karampelis / Postdoctoral Fellow
Phone Number
+30 210 9656310 (ext. 203)
Email
Address
BSRC "Alexander Fleming"
34 Fleming Street, 16672
Vari, Greece
 |
Unit development is supported by: the Hellenic Foundation for Research & Innovation (HFRI) ELIDEK; 1st Call for H.F.R.I. Research Projects to Support Faculty Members & Researchers and Procure High-Value Research Equipment (Grant ID: 3780) to G. Kollias. |