Science Projects
The Politis Lab and its associated projects focus on two primary themes:
1. Advancing technology to enable the investigation of challenging protein samples.
2. Applying these technologies to uncover molecular mechanisms underlying disease-related states
1. Technology development
The Politis Lab has pioneered HDX-MS (Hydrogen-Deuterium Exchange Mass Spectrometry) strategies to study membrane proteins—such as transporters and G-Protein Coupled Receptors (GPCRs)—within their native lipid environments. We have demonstrated that HDX-MS can serve as a drug profiling tool, capable of monitoring ligand binding to GPCRs and classifying them based on observed allosteric effects (Toporowska et al., Nature Communications, 2024).
Despite our leadership in this field, challenges remain—particularly in analyzing integral membrane proteins (IMPs) due to lipid-induced ion suppression and chromatographic interference. To address this, we aim to develop in situ HDX-MS methods that capture the conformational dynamics of membrane proteins in their native settings.
Liposomes offer the advantages of other membrane mimetics—such as the ability to fine-tune lipid composition—while also introducing features like membrane curvature, lateral pressure, and protein directionality that more closely mimic native biological membranes. Due to their large size, liposomes are particularly useful for studying multicomponent systems. Their aqueous lumen not only supports the maintenance of membrane potential but also enables the encapsulation of activity reporter systems.
Despite these advantages, the application of liposomes in mass spectrometry (MS) has largely been limited to small peptides or single-pass transmembrane proteins. This limitation arises from the challenges associated with handling and analyzing heterogeneous samples using current methodologies.
In our group, we build upon recent advances in managing sample heterogeneity to generate liposomes with single protein orientation and high reproducibility. Furthermore, we leverage the positive-inside rule to ensure correct protein insertion into native membranes. This controlled directionality not only facilitates the application of energy gradients but also mitigates the risk of misinterpreting kinetics in hydrogen-deuterium exchange mass spectrometry (HDX-MS), thereby enhancing data quality.
Native Membranes. While liposomes offer a simplified model system, isolated membrane vesicles provide a more physiologically relevant environment for studying membrane proteins (MPs) within native lipid bilayers. These vesicles retain key features such as membrane-associated protein interaction partners and components that maintain electrochemical gradients. Additionally, they preserve lipid asymmetry, a feature that is difficult to replicate in synthetic systems.
To study MPs in this native context, we employ a delipidation strategy to fingerprint overexpressed proteins from E. coli. We then isolate the inner membranes and reconstitute them into lipid vesicles. Protein interactions are subsequently analyzed in situ using hydrogen-deuterium exchange mass spectrometry (HDX-MS).
2. Applications
Transporters
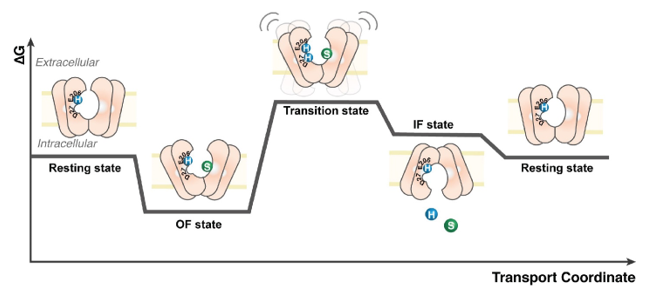
Transporters are essential biomolecules that mediate the movement of substrates across biological membranes. However, obtaining structural information about their conformational states remains challenging, particularly for eukaryotic systems. These difficulties arise from their intrinsic dynamics and the low yield of protein purification using traditional structural biology techniques.
To address these challenges, we employ hydrogen-deuterium exchange mass spectrometry (HDX-MS) to capture the conformational landscapes of sugar transporters such as XylE and LacY, as well as their human homologue, GLUT1. Our work has elucidated how the lipid environment modulates the conformational dynamics of these transporters (Martens et al., Nature Communications. 2018; Martens et al. Nature protocols 2019; Jia et al. Nature Communications, 2020;).
Building on these insights, we developed a comprehensive HDX-MS protocol optimized for experiments in lipid nanodiscs, which we integrate with molecular dynamics simulations (Martens et al. Nature protocols 2019). This approach has revealed how ligand binding influences structural dynamics and has clarified the role of lipids in regulating the oligomeric states of eukaryotic transporters.
These findings are detailed in our study (Pyle et al., Cell Chemical Biology, 2018), which provides a mechanistic framework for understanding transporter behavior in native-like environments.
G Protein-Coupled Receptors (GPCRs)
Probing the conformational dynamics of GPCRs in native environment
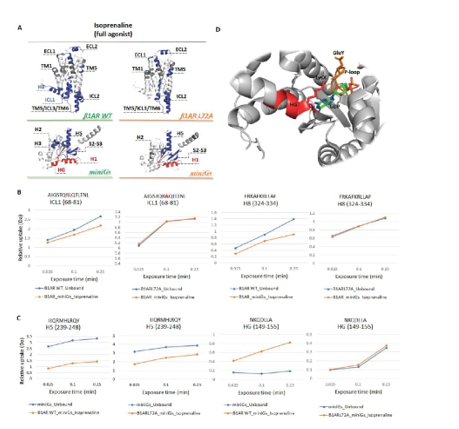
Hydrogen-deuterium exchange mass spectrometry (HDX-MS) is a highly sensitive technique for probing the structural dynamics of membrane proteins under near-native conditions. Our group has recently developed protocols to monitor conformational changes in eukaryotic proteins, including G protein-coupled receptors (GPCRs). Using HDX-MS, we characterized the structural dynamics of the β1-adrenergic receptor (β1AR) in complex with nine ligands representing different pharmacological classes—agonists, partial agonists, and antagonists (Yen et al., Nature Chemistry, 2022; Toporowska et al., Nature Communications, 2024).
Our analysis revealed that ligand-induced dynamic signatures across the GPCR structure cluster according to ligand modality. Notably, we observed consistent destabilization of intracellular loop 1 (ICL1) upon full agonist binding, whereas antagonist binding led to its stabilization. These findings suggest that increased flexibility in ICL1 may be a critical feature facilitating G-protein recruitment.
We collaborate with the Skretas Lab to investigate the conformational dynamics of membrane proteins (MPs) in their native environments. The Skretas Lab has developed a novel set of low-toxicity E. coli strains—SuptoxD, SuptoxR, and their derivatives—for enhanced production of recombinant MPs (Michou et al., ACS Synthetic Biology, 2019). These strains significantly increase the volumetric yield of well-folded MPs from both prokaryotic and eukaryotic sources, including the neurotensin receptor 1 (NTSR1).
To study the structural dynamics of NTSR1—a G protein-coupled receptor (GPCR) involved in regulating blood pressure, analgesia, and thermoregulation—we employ hydrogen-deuterium exchange mass spectrometry (HDX-MS). This highly sensitive technique monitors hydrogen exchange with deuterium to reveal conformational changes.
We specifically examine how binding of the lipid PIP2 influences NTSR1 dynamics and induces allosteric effects. Using the specialized E. coli strains, we also investigate lipid-mediated modulation in other eukaryotic MPs. Furthermore, we study MPs in situ by solubilizing them in lipid vesicles derived directly from inner membranes.
DISEASE STATES
Inhibitory mechanism and molecular interactions of the hypoxia upregulated protein 1
The Kollias Lab is internationally recognized for its expertise in elucidating the molecular and cellular mechanisms underlying chronic inflammatory disorders, including Crohn’s disease and rheumatoid arthritis. The lab has developed innovative pipelines for cheminformatics and in vitro testing, alongside conducting preclinical evaluations in mouse models of chronic inflammation.
In this study, we investigate – in collaboration with the Kollias Lab - the molecular and conformational fingerprints of hypoxia upregulated protein 1 (HYOU1), an endoplasmic reticulum-resident chaperone implicated in tumorigenesis and other pathological conditions (Tsukamoto et al., J Clin Invest, 1996). Our aim is to characterize the binding interactions of HYOU1 with small molecules such as ATP and other known or potential therapeutic compounds.
To achieve this, we will employ a combination of hydrogen-deuterium exchange mass spectrometry (HDX-MS) to probe dynamic structural changes upon ligand binding, and native mass spectrometry (native MS) to assess binding stoichiometry and affinities. These experiments will not only provide novel insights into the molecular partners and mechanisms of HYOU1 but also serve as a proof-of-concept for our broader drug discovery platform.
Structural characterization of mitochondrial solute carrier SLC25
SLC25A46 is a recently identified mitochondrial outer membrane protein implicated in a broad spectrum of rare neurological disorders, including optic atrophy, Leigh syndrome, and progressive myoclonic ataxia. It belongs to the Solute Carrier 25 (SLC25) family of nuclear-encoded mitochondrial transporters and plays a critical role in mitochondrial dynamics and cristae architecture.
Douni’s group previously developed a neurological mouse model harboring a nonsense mutation in the Slc25a46 gene (Terzenidou et al, PloS genetics, 2017). These mutant mice recapitulate key clinical features observed in patients, such as ataxia, optic atrophy, and cerebellar hypoplasia—phenotypes that can be rescued by expression of the human ortholog. Histopathological analysis further revealed novel lesions, including disrupted cerebellar and retinal cytoarchitecture, as well as abnormalities at the neuromuscular junction.
This mouse model provides a valuable platform for dissecting the molecular mechanisms underlying SLC25A46-associated pathologies. In this study, we will employ hydrogen-deuterium exchange mass spectrometry (HDX-MS) to investigate conformational dynamics associated with pathogenic mutations. In parallel, we will use cryo-electron microscopy (cryo-EM) and computational modeling (in collaboration with Kastritis) to resolve the structures of both apo and mutant forms of SLC25A46.
Additionally, we will analyze the protein in outer membrane vesicles to better understand its function in a native-like environment. The integration of HDX-MS, cryo-EM, and computational tools will not only enable the construction of a disease-relevant structural model but also establish a framework for studying similar mitochondrial systems.