recoup: remastering next generation sequencing signal visualization
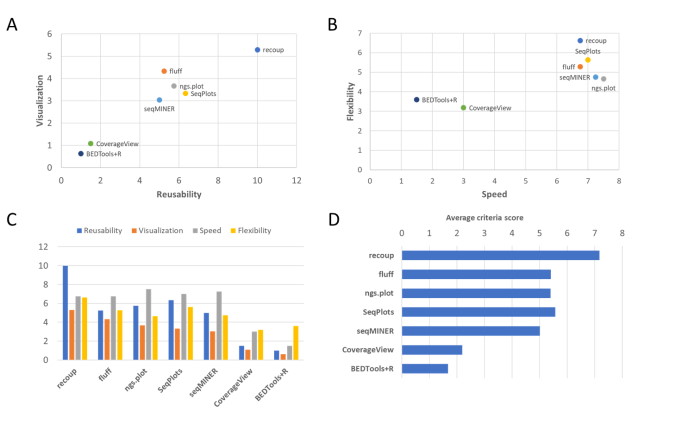
recoup, a new software package by the Moulos lab, published in BMC Bioinformatics, allows for comprehensive and versatile visualization of summarized NGS signals. Backed by a small user-study, recoup’s purpose spans applications from routine quality control and visual hypothesis checks up to publication quality figures. [Pubmed], [DOI]
The continuing emergence of new genomic sequencing protocols and the resulting generation of ever larger datasets continue to challenge the meaningful summarization and visualization of the underlying signal generated to answer important qualitative and quantitative biological questions. While existing solutions for the comprehensive visualization of NGS data signal offer satisfying results, they are often compromised regarding issues such as effortless tracking of processing and preparation steps under a common computational environment, visualization quality and user friendliness.
To tackle such issues as well as address modern data visualization needs, the Moulos lab has developed recoup, a Bioconductor package for the quick, flexible, versatile, and accurate visualization of genomic coverage profiles generated from Next Generation Sequencing data. Coupled with a database of precalculated genomic regions for multiple organisms, recoup offers processing mechanisms for quick, efficient, and multi-level data interrogation with minimal effort, while at the same time creating publication-quality visualizations. Special focus is given on plot reusability, reproducibility, and real-time exploration and formatting options, operations rarely supported in similar visualization tools in a profound way. recoup was assessed and compared to other solutions during a small user-study and using several qualitative user metrics. It was found to balance the tradeoff between important package features, including speed, visualization quality, overall friendliness, and the reusability of the results with minimal additional calculations.