06/02/2020
A collaborative study involving advanced bioinformatics analyses conducted by Panagiotis Moulos, Stavros Niarchos researcher at BSRC Alexander Fleming and Iannis Talianidis lab at IMBB-FORTH provides evidence of chromatin bookmarking by the non-pioneer transcription factors C/EBPα and HNF4α which then is key to future gene activation during liver development. [Pubmed]
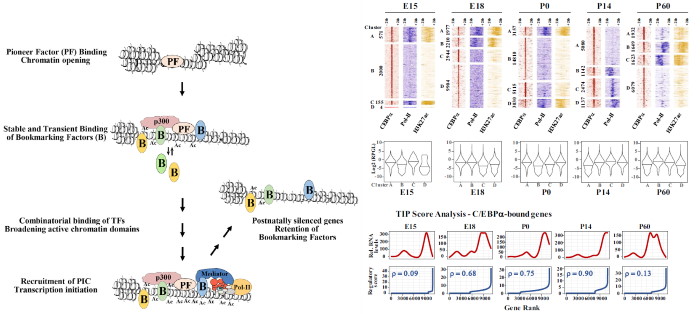
Transcription factor binding to enhancer and promoter regions critical for homeostatic adult gene activation is established during development. To understand how cell-specific gene expression patterns are generated, the researchers studied the developmental timing of association of two prominent hepatic transcription factors with gene regulatory regions. Strikingly, one of the main findings was that transcription factor binding dynamics poorly correlate with gene activation timing when measured in respective time course experiments in key developmental stages of the mouse, as revealed by correlation with H3K27ac and RNA polymerase signals as well as gene expression profiling. Instead, gene activation often occurred long after the first detectable recruitment of C/EBPα or HNF4α, suggesting a bookmarking mechanism which destabilizes condensed chromatin to provide access for other transcription factors activating transcription. Overall, the temporal dynamics of transcription factor binding uncovered in the study suggest that gene activation mechanisms during development, cell-cycle progression, and evolution are shaped by common regulatory principles.
KEYWORDS: CEBP; HNF4; chromatin; development; gene regulation; liver; transcription factor
The study was published in the CELL Reports Volume 30, Issue 5, 4 February 2020, Pages 1319-1328.e6; DOI: 10.1016/j.celrep.2020.01.006