02/09/2019
Highlights
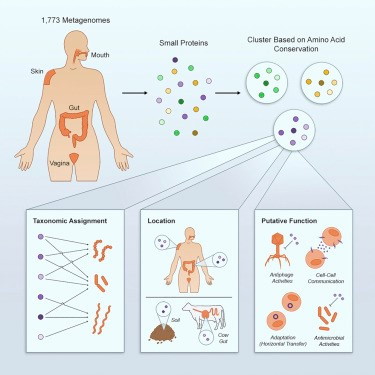
• A genomic approach finds >4,000 conserved small proteins in human microbiomes
• Most of these proteins have no known function or domain
• A database provides insights into potential function of these proteins
• Over 30% of the small proteins are predicted to be involved in cell-cell communication
Small proteins (fewer than 50 amino acids in length) are traditionally overlooked due to computational and experimental difficulties in detecting them. To systematically identify small proteins, scientists carried out a comparative genomics study on 1,773 human-associated metagenomes from four different body sites. They describe >4,000 conserved protein families, the majority of which are novel. ∼30% of these protein families are predicted to be secreted or transmembrane, thus being involved in cell-cell communication. Over 90% of the identified small protein families have no known domain and almost half of them are not represented in any reference genome. The team additionally identified putative housekeeping, mammalian-specific, defense-related proteins and protein families that are likely to be products of horizontal transfer events.
Overall, the study suggests that small proteins are highly abundant and particularly those of the human microbiome may perform diverse functions that have not been previously reported.
Τhe Pavlopoulos team crucially contributed in the collaborative study lead by Dr. Ami Bhatts from Stanford University School of Medicine, while One Codex, DOE Joint Genome Institute and Lawrence Berkeley National Laboratory also participated.
The study was published in Cell journal, Vol. 178, Issue 5, p1245-1259.E14, August 08 2019 DOI: https://doi.org/10.1016/j.cell.2019.07.016
PubMed: https://www.ncbi.nlm.nih.gov/pubmed/?term=31402174
Other links: http://med.stanford.edu/news/all-news/2019/08/human-microbiome-churns-out-thousands-of-tiny-novel-proteins.html