01/09/2020
A new study by the Moulos lab reveals that combining RNA-Seq analysis algorithms not only improves accuracy, but shows robustness against different normalizations, alleviates downstream biases including gene-length bias in pathway analysis and improves long non-coding RNAs expression analysis. [Pubmed]
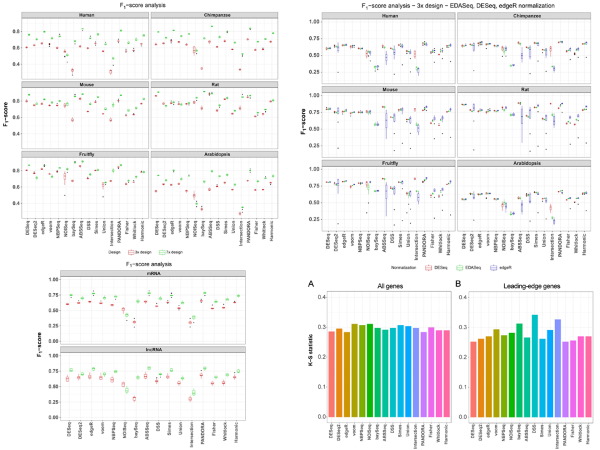
The investigation of differential gene expression patterns through RNA-Seq comprises a routine task in the daily lives of molecular bioscientists, who produce vast amounts of data requiring proper management and analysis. Despite widespread use, there are still no widely accepted golden standards for the normalization and statistical analysis of RNA-Seq data. A new study by the Moulos lab significantly extends the previously introduced PANDORA algorithms for the analysis of RNA-Seq data (https://doi.org/10.1093/nar/gku1273) and presents the redesigned underlying Bioconductor package metaseqR2 (https://bioconductor.org/packages/release/bioc/html/metaseqR2.html).
The study builds upon previous work and reveals that the systematic combination of high performant algorithms for the statistical analysis of RNA-Seq data, not only improves the overall accuracy by optimizing the tradeoff between true and false hits, but also demonstrates notable robustness against different normalization frameworks and alleviates further downstream analysis biases including the gene-length bias propagation in pathway analysis and the differential expression of long non-coding RNAs. Furthermore, it introduces the functionalities of metaseqR2, a new Bioconductor package that hosts the PANDORA algorithm, and interfaces to several popular differential expression algorithms, normalization frameworks and p-value combination methods, as well as the completely redesigned, re-engineered and fully interactive analysis report.