Genome integrity is essential for accurate gene expression and epigenetic inheritance. Our research aims to deepen our understanding of the underlying regulatory mechanisms, whose deviation may lead to rare neurodevelopmental progeroid disorders and/or cancer.
Main scientific directions
Our scientific directions during this period include:
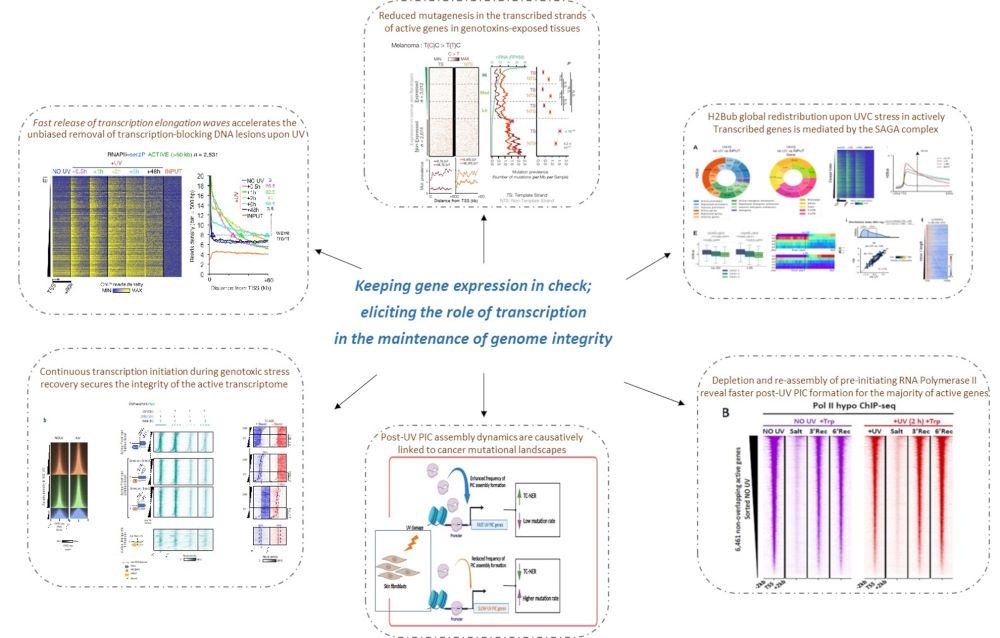
1. Deciphering the role of transcription in the maintenance of genome integrity
Our studies revealed a multi-layered transcriptional response and associated chromatin dynamics that are triggered in human skin fibroblasts upon exposure to genotoxic stress such as UV irradiation, cigarette smoke, and several chemotherapeutics currently in clinical use. This response relies on active and continuous transcription initiation and is associated with chromatin remodelling events of genes and regulatory regions including promoters, PROMPTS, and enhancers. Notably, this mechanism promotes genetic accuracy by ensuring fast and efficient repair of the active transcriptome and results in limited mutagenesis of genotoxic-agent exposed tissues, such as skin and lungs.
Relevant publications:
1. Liakos Anastasios*, Anna-Chloe Synacheri#, Dimitris Konstantopoulos#, Stefos Georgios, Matthieu D. Lavigne, Maria Fousteri*. Enhanced frequency of transcription pre-initiation complexes assembly after exposure to UV irradiation results in increased repair activity and reduced probabilities for mutagenesis. Nucleic Acids Research, gkad593, https://doi.org/10.1093/nar/gkad593
2. Fanourgakis Smaragda#, Anna-Chloe Synacheri#, Matthieu D. Lavigne, Dimitris Konstantopoulos*, Maria Fousteri*. 2023, ‘Histone H2Bub1 dynamics in the 5' region of active genes are tightly linked to the UV-induced transcriptional response’. Computational & Structural Biotechnology Journal, 21:614-629. doi 10.1016/j.csbj.2022.12.013
3. Liakos A, Konstantopoulos D, Lavigne MD*, Fousteri M*. 2020, ‘Continuous transcription initiation guarantees robust repair of all transcribed genes and regulatory regions’. Nature Communications 11 (1): 916. DOI: 10.1038/s41467-020-14566-9.
4. Lavigne MD, Konstantopoulos D, Ntakou-Zamplara KZ, Liakos A, Fousteri M. 2017, Global unleashing of transcription elongation waves in response to genotoxic stress restricts somatic mutation rate. Nature Communications 8(1): 2076. doi: 10.1038/s41467-017-02145-4.
2. aniFOUND; Analysing the associated proteome and genomic landscape of the repaired nascent non-replicative chromatin
We have developed aniFOUND, a high-throughput antibody-free methodology that can integrate proteome analysis (aniFOUND-MS) and genome-wide mapping (aniFOUND-seq) to characterise the repaired nascent chromatin landscape throughout the genome. As a case study, we used UVC-induced DNA lesions that were repaired by nucleotide excision repair (NER). Our findings underscore the efficacy of aniFOUND to elucidate mechanisms of skin carcinogenesis and to interpret the aetiology of NER-related rare disorders.
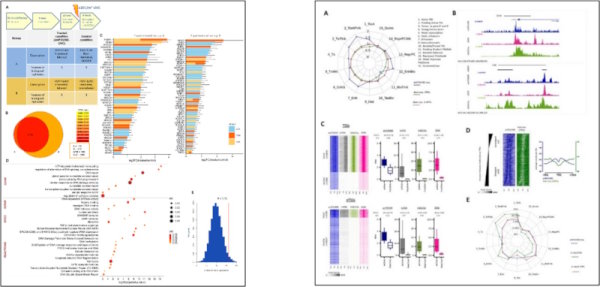
With aniFOUND-MS, we revealed previously unknown players involved in UV-triggered DNA Damage responses and the restoration of the damaged chromatin.
We also established a novel protocol for deep sequencing of the repaired nascent DNA segments (aniFOUND-seq) and specific analysis pipelines and we assessed UV-induced unscheduled (non-replicative) DNA synthesis in chromosomal regions with different features, providing unbiased spatio-temporal insights into genome maintenance mechanisms.
Relevant publication:
Georgios C Stefos, Eszter Szantai, Dimitris Konstantopoulos, Martina Samiotaki, Maria Fousteri, 2021. “aniFOUND: analysing the associated proteome and genomic landscape of the repaired nascent non-replicative chromatin”. Nucleic Acids Research, 49 (11): e64; gkab144, https://doi.org/10.1093/nar/gkab144
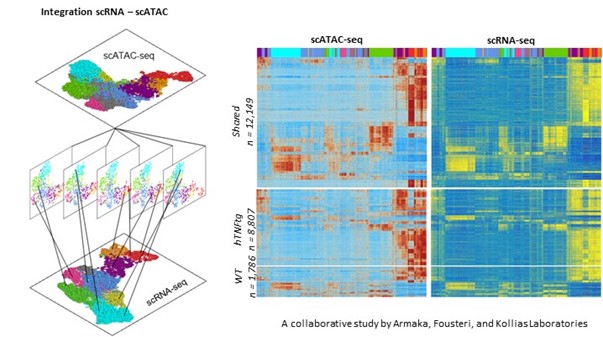
3. Investigation of the molecular paths, GRNs and cellular heterogeneity that underlie disease-driven biological processes by single-cell (sc) NGS (Next Generation Sequencing) approaches.
Employing scRNA-seq and scATAC-seq methodologies and integrative analyses we recently generated and successfully resolved functional sc maps of synovial fibroblasts (SF) populations derived from naïve and hTNFtg mice (mice that overexpress human TNF, a murine model for Rheumatoid Arthritis). Our studies defined a dynamic SF landscape from health to arthritis, highlighting the underlying molecular switches, spatiotemporal dynamics, and gene regulatory networks and providing novel insights into the pathogenic remodeling of synovial microenvironment.
Relevant publication:
Marietta Armaka#*, Dimitris Konstantopoulos#, Christos Tzaferis#, Matthieu D Lavigne#, Maria Sakkou#, Anastasios Liakos, Petros P Sfikakis, Meletios A Dimopoulos, Maria Fousteri*, George Kollias*, 2022. ‘Single-cell multimodal analysis identifies common regulatory programs in Synovial Fibroblasts of Rheumatoid Arthritis patients and modelled TNF-driven arthritis’. Genome Medicine, doi:
https://genomemedicine.biomedcentral.com/articles/10.1186/s13073-022-01081-3
# equally contributed, first authors
*Corresponding authors
4. Elucidating disorders of transcription de-regulation that are associated with severe neurodevelopmental abnormalities and premature aging.
In the current project, we aim to shed light on the molecular aetiology of the progeroid disorder Cockayne Syndrome that is characterised by severe neurodegenerative and developmental abnormalities.
Relevant publications:
1. Liakos A, Lavigne MD, Fousteri M., 2017. Personalized medicine, Book chapter on “Nucleotide Excision Repair: Lessons from Neurodegeneration to Cancer”. Springer Nature, Adv. Exp. Med.Biology; 1007:17-39, doi:10.1007/978-3-319-607337_2.
2. Zoi Spyropoulou, Angelos Papaspyropoulos, Nefeli Lagopati, Vassilios Myrianthopoulos, Alexandros G. Georgakilas, Maria Fousteri, Athanassios Kotsinas and Vassilis G. Gorgoulis (2021). ‘Cockayne Syndrome Group B (CSB): The Regulatory Framework Governing the Multifunctional Protein and Its Plausible Role in Cancer’. Cells, https://doi.org/10.3390/cells10040866