Genome Architecture in one and three dimensions
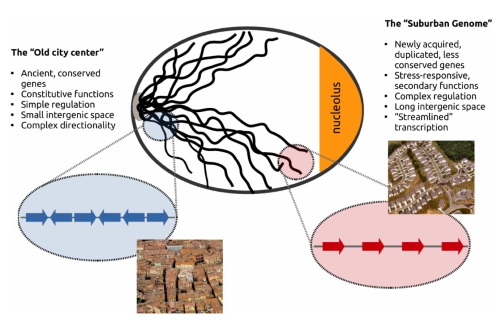
We are viewing genomes as architectures, that is as complex systems, whose functional parts evolve under certain constraints that are related to the confines of space and functionality. We are interested in understanding the relationships between gene regulation and the genes' spacing, relative proximity and distances in one and the three dimensions. In this context we explore spatial models that are inspired from statistical physics or the social sciences in systems approaches that deal with the genomes as evolving "Landscapes".
[Selected Works]
•Tsochatzidou M, Malliarou M, Papanikolaou N, Roca J and Nikolaou C. (2017). Genome urbanization: Clusters of topologically co-regulated genes delineate functional compartments in the genome of S. cerevisiae. Nucleic Acids Research, 45 (10), 5818-5828. https://dx.doi.org/10.1093/nar/gkx198
•Nikolaou C (2018) Invisible Cities: Segregated Domains in the yeast genome with distinct structural and functional attributes. Current Genetics, 64 (1), 247-258. https://dx.doi.org/10.1007/s00294-017-0731-6
•Mavropoulos Papoudas S, Papanikolaou N and Nikolaou C., (2020) Monitoring the prolonged TNF stimulation in space and time with topological-functional networks. Computational and Structural Biotechnology Journal, 18, 220-229. https://doi.org/10.1016/j.csbj
•Salataj E, Stathopoulou C, Hafþórsson RA, Nikolaou C and Spilianakis GC (2019) Developmental Conservation of microRNA Gene Localization at the Nuclear Periphery. PLoS One, 14(11):e0223759. https://doi.org/10.1371/journal.pone.0223759.
Genome Structure and Epigenomics in disease
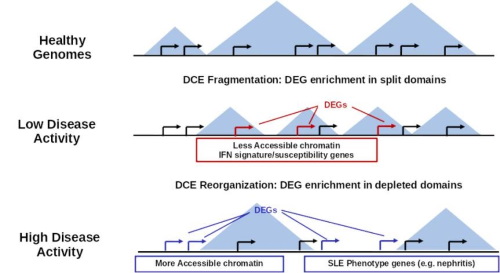
Complex inflammatory diseases and cancer are prime examples of large-scale gene de-regulation. In this context we are interested in understanding: a) the impact of chromatin and epigenetic factors on disease progression and b) the succession of regulatory events that may help us identify the driving agents of pathology.
[Selected Works]
•Roulis M, Nikolaou C, Kotsaki E, Kaffe E, Karagianni N, Koliaraki V, Salpea K, Ragoussis J, Aidinis V, Martini E, Becker C, Herschman HR, Vetrano S, Danese S and Kollias G (2014) Intestinal myofibroblast-specific Tpl2-Cox-2-PGE2 pathway links innate sensing to epithelial homeostasis. Proceedings of the National Academy of Sciences, U.S.A. 111 (43): E4658-E4667 http://dx.doi.org/10.1073/pnas.1415762111
•Ntougkos E, Chouvardas P, Roumelioti F, Ospelt C, Frank-Bertoncelj M, Fihler A, Buckley CD, Gay S, Nikolaou C# and Kollias G# (2017) Genomic responses of mouse synovial fibroblasts during TNF-driven arthritogenesis greatly mimic those of human rheumatoid arthritis. Arthritis and Rheumatology, 69 (8): 1588-1600. https://dx.doi.org/10.1002/art.40128
•Ntasis VF, Panousis NI, Tektonidou MG, Dermitzakis ET, Boumpas DT, Bertsias GK, Nikolaou C (2020) Extensive fragmentation and re-organization of gene co-expression patterns underlie the progression of Systemic Lupus Erythematosus https://doi.org/10.1101/2020.01.28.922559
•Papadaki C, Stratigos M, Markakis G, Spiliotaki M, Mastrostamatis G, Nikolaou C, Mavroudis D and Agelaki S (2018) Circulating microRNAs in the early prediction of disease recurrence in primary breast cancer. Breast Cancer Research. 201820:72 https://doi.org/10.1186/s13058-018-1001-3
Nucleosome Positioning and regulation of gene expression

We are developing methods to assess the local nucleosome structure around transcription initiation and splicing sites and employ them in the analysis of gene expression data in a chromatin context. Our work on exons being defined at chromatin level has led to the discovery of co-transcriptional splicing. We have also contributed in the elaboration of models for the statistical positioning of nucleosomes in eukaryotes.
[Selected Works]
•Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcárcel J and Guigo R. (2009) Nucleosome positioning as a determinant of exon recognition. Nature Structural & Molecular Biology, 16(9):996-1001. http://dx.doi.org/10.1038/nsmb.1658
•Nikolaou C, Althammer S, Beato M and Guigo R. (2010) Structural constraints revealed in consistent nucleosome positions in the genome of S. cerevisiae. Epigenetics and Chromatin 3 (1) 20 http://dx.doi.org/10.1186/1756-8935-3-20
•Nikolaou C, Bermudez I, Manichanh C, García-Martinez J, Guigo R, Perez-Ortin JE and Roca J (2013) Topoisomerase II regulates yeast genes with singular chromatin architectures. Nucleic Acids Research 41 (20): 9243-9256 http://dx.doi.org/10.1093/nar/gkt707
Modeling Genome Regulation during development
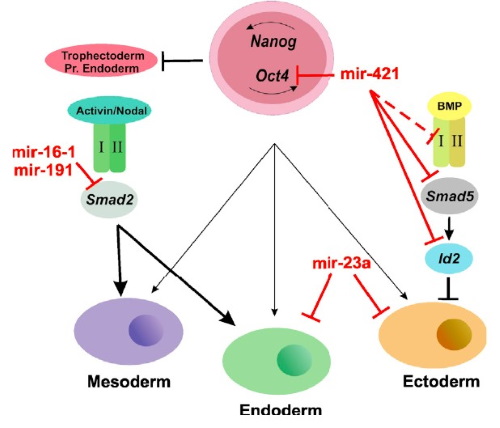
We are particularly interested in monitoring gene regulation dynamics in time. In this respect we have extensive work on developmental system, in which we focus on time-course dependent data during differentiation processes. Our involvement is centered on the analysis of time-course expression and regulation data and the reconstruction of the Gene Regulatory Networks (GRN).
[Selected Works]
•Hadjimichael C, Nikolaou C, Papamatheakis J and Kretsovali A. (2016) MicroRNAs for Fine-Tuning of Mouse Embryonic Stem Cell Fate Decision through Regulation of TGF-β Signaling. Stem Cell Reports 6 (3): 292-301 http://dx.doi.org/10.1016/j.stemcr.2016.01.004
•Hadjimichael C, Chanoumidou K, Nikolaou C, Klonizakis A, Theodosi GI, Makatounakis T, Papamatheakis J and Kretsovali A. (2017) Promyelocytic Leukemia (PML) protein is an essential regulator of stem cell pluripotency and somatic cell reprogramming. Stem Cell Reports, 8 (5), 1366-1378 http://dx.doi.org/10.1016/j.stemcr.2017.03.006
•Chanoumidou K, Hadjimichael C, Athanasouli P, Ahlenius H, Klonizakis A, Nikolaou C, Drakos E, Kostouros A, Stratidaki I, Grigoriou M, Kretsovali A. (2018) Groucho related gene 5 (GRG5) is involved in embryonic and neural stem cell state decisions. Sci Rep. 2018 Sep 13;8(1):13790. https://doi.org/10.1038/s41598-018-31696-9
•Sachini N, Arampatzi P, Klonizakis A, Nikolaou C, Makatounakis T, Lam E W-F., Kretsovali A and Papamatheakis J (2019) Promyelocytic leukemia protein (PML) controls breast cancer cell proliferation by modulating Forkhead transcription factors. Molecular Oncology, 13, 1369-1387. https://doi.org/10.1002/1878-0261.12486
-Omics Data Analysis and Machine Learning

We employ computational approaches in the analysis of complex multi-omics data with focus on transcriptomic analyses. We build computational pipelines for the reconstruction of Gene Regulatory hierarchies, the detection of gene biomarkers through gene prioritization and the association of microRNAs with potential gene targets.
[Selected Works]
•Papadaki C, Monastirioti A, Rounis K, Makrakis D, Kalbakis K, Nikolaou C, Mavroudis D and Agelaki S, (2020) Circulating MicroRNAs Regulating DNA Damage Response and Responsiveness to Cisplatin in the Prognosis of Patients with Non-Small Cell Lung Cancer Treated with First-Line Platinum Chemotherapy. Cancers 2020, 12(5), 1282. https://doi.org/10.3390/cancers12051282
•Karagianni N, Kranidioti K, Fikas N, Tsochatzidou M, Chouvardas P, Denis MC, Kollias G and Nikolaou C (2019) An integrative transcriptome analysis framework for drug efficacy and similarity reveals drug-specific signatures of anti-TNF treatment in a mouse model of inflammatory polyarthritis. PLoS Computational Biology 15(5): e1006933. https://doi.org/10.1371/journal.pcbi.1006933
•Chouvardas P, Kollias G and Nikolaou C. (2016) Inferring active regulatory networks from gene expression data using a combination of prior knowledge and enrichment analysis. BMC Bioinformatics 17(5):181 http://dx.doi.org/10.1186/s12859-016-1040-7
Genomes as complex systems
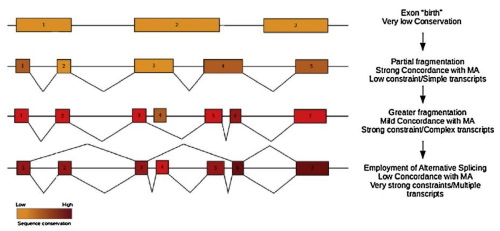
We have been studying various statistical aspects of genomic sequences and genome organization from the early 2000s, primarily focusing on the evolution of gene structure and gene distribution in relation to the process of DNA replication. We maintain an active line of research centered on aspects of exon evolution, gene spacing and transcription directionality with the aim of understanding how genomes evolve in time.
[Selected Works]
•Nikolaou C. (2014) Menzerath–Altmann law in mammalian exons reflects the dynamics of gene structure evolution. Computational Biology and Chemistry, 53: 134-143. http://dx.doi.org/10.1016/j.compbiolchem.2014.08.018
•Nikolaou C and Almirantis Y (2005) A Study on the correlation of nucleotide skews and the positioning of the Origin of Replication. Different modes of replication in bacterial species. Nucleic Acids Research, 33, 6816-6822. https://doi.org/10.1093/nar/gki988
•Nikolaou C and Almirantis Y (2006) Deviations from Chargaff's second parity rule in organellar DNA Insights into the evolution of organellar genomes. Gene , 381:34-41. http://dx.doi.org/10.1016/j.gene.2006.06.010