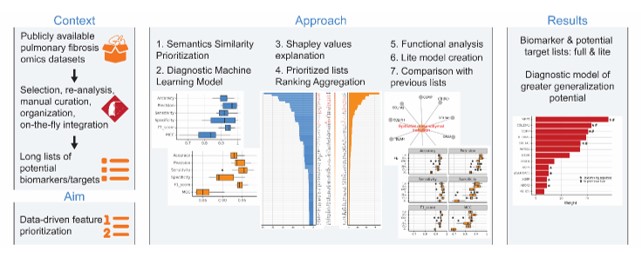
Exploiting the vast transcriptomic information stored at Fibromine, all publicly available omics datasets on Idiopathic Pulmonary Fibrosis (IPF) were compared, and deregulated genes in most datasets were prioritized via machine learning, resulting in a list of genes with highly predictive statistical power.
Idiopathic pulmonary fibrosis (IPF) is an incurable pathology of unknown etiology with reported increasing prevalence and alarming association with COVID-19. State of the art anti-fibrotic agents, nintedanib and pirfenidone, can only retard disease progression in exchange for often serious side effects. In an attempt to ameliorate the current IPF treatment status, novel disease targets/biomarkers should urgently be discovered. In a previous publication of the lab, Fanidis et al. (2021) (PMID: 34741074) selected, manually curated and consistently reanalyzed top quality publicly available IPF-related omics datasets. The results were organized in a database called Fibromine and can be accessed, mined and on-the-fly combined via various functionalities offered via the homonym Shiny application. To exploit the aforementioned centralized data source, in their most recent publication, Fanidis et al. (2023) (PMID: 37007651) have crafted an ensemble machine learning classifier to separate IPF from control phenotypes based on Fibromine-hosted gene expression data. Shapley values were used for model interpretation enabling the formation of two novel biomarker lists. Both are characterized by an at least equal classification performance compared to that of previously published biomarker sets. Examination of the proposed feature sets revealed both well-established as well as latent deregulated genes that can be used, among others, as a target pool for future wet lab experimentations.
[PubMed]