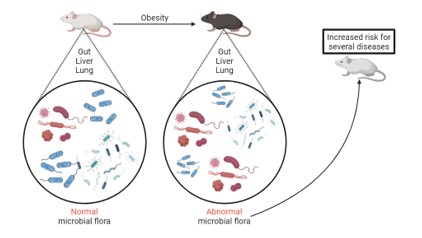
Enabling the study of microbiomics locally, a 16S rRNA deep sequencing pipeline has been created and was used to examine and compare the microbial communities of the gut, liver, and lung of obese and lean mice.
Microbiome is increasingly recognized as a major player in tissue homeostasis in health and disease, modulating a variety of host functions, including immunity and inflammation, as well as energy homeostasis and metabolism. In this work, we examined the microbial composition of the guts, livers, and lungs of mice fed a high-fat diet (HFD) compared to littermates fed a matched control diet (CD). Towards this goal, we employed sequencing of seven 16S rRNA gene hypervariable regions. As shown, comparison of the local microbiomes indicated that lung tissue has the least diverse microbiome under healthy conditions, while microbial diversity in the healthy liver clustered closer to the gut. Obesity was shown to increase microbial complexity along the gut-liver-lung axis. Although the lung is the tissue with the least diverse microbiome under healthy conditions, its microbial diversity was affected the most by HFD-driven obesity and reached levels similar to those recorded for the other two tissues. In healthy CD mice, all three tissues examined (gut, liver, and lung) were populated chiefly by four phyla: Proteobacteria, Actinobacteria, Bacteroidetes, and Firmicutes. HFD-driven obesity suppressed Bacteroidetes in the gut, while it increased the diversity of Firmicutes in all three tissues as was observed for the very first time. Among Firmicutes, an abundance of Staphylococcus was characterized by an increasing tendency in all three tissues upon obesity. In conclusion, in addition to lipid deposition throughout the body and the triggering of NAFLD, obesity was shown to increase microbial complexity along the gut-liver-lung axis and, as a result, to possibly predispose mice to a series of metabolic diseases via increased abundance in Staphylococcus and other species.
[PubMed]