Fibromine, a new database and data mining tool for target discovery in pulmonary fibrosis has been created to explore the numerous related publicly available datasets.
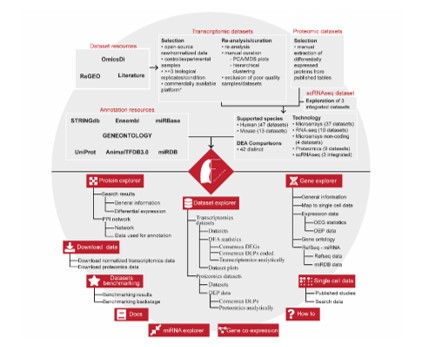
Idiopathic pulmonary fibrosis is a lethal pathology usually affecting elder individuals with a smoking history. Despite intensive efforts, research community has not yet discovered neither its origin nor any curative option. Given its increasing prevalence, proposal of new disease targets is of vital importance. Towards that goal, in silico analyses of publicly available datasets can accelerate any research attempt by inexpensively examining a huge volume of samples. Nevertheless, such endeavor requires a significant level of computational expertise and even then, datasets collection and curation do necessitate a huge number of working hours. To address the above limitations in the exploration of wealthy IPF high-throughput datasets, Fanidis et al. (2021) (PMID: 34741074) have created Fibromine, a centralized database of IPF-related transcriptomics and proteomics data accessible via the homonym user-friendly web application. In Fibromine, carefully selected, manually curated and extensively re-analyzed datasets of both human and mouse origin can be mined and combined in real time. In addition, gene-specific expression patterns can be interrogated at both bulk and single cell levels, while several other functionalities support investigation of miRNA-mRNA interactions and gene co-expression phenomena. Moreover, protein explorer opens a window to the world of differential protein abundance and offers the opportunity of exploring protein-protein interactions via the creation of pathology-specific interaction networks. In conclusion, Fibromine aims to acceleration of IPF research for novel disease-implicated features by democratizing the use of omics data.
[PubMed]